Predicting genotypic evolution
To combat pathogens, agricultural pests, and invasive species we must understand how phenotypic changes, such as resistance, arise from genotypic dynamics. We aim to build and validate models of genotypic evolution from the ground up.
The genotype-phenotype map
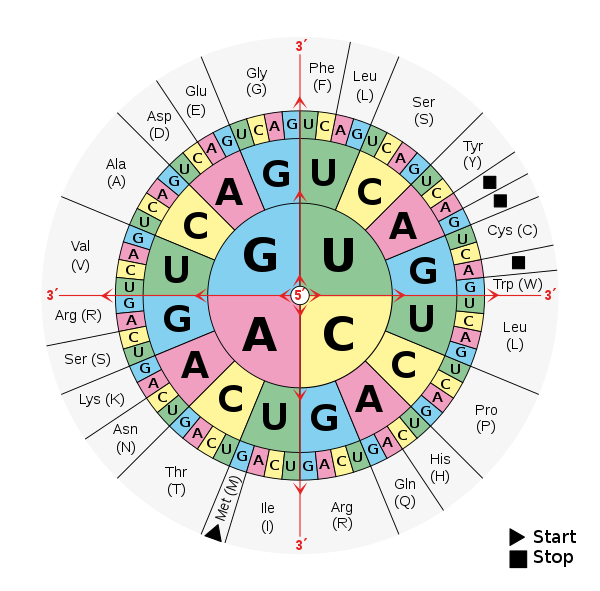
The mapping between nucleotides and amino acids one of the most fundamental components of our understanding of the evolution of life. Each amino acid is determined by a sequence of three nucleotides: the codon. The triplicate nature of the codon was first revealed by the Crick, Brenner et al. experiment (1961). Nucleotide substitutions in a gene change the amino acids translated at the ribosome and thus the chemical properties of the proteins the gene encodes.
Markovian jump process for nucleotide substitutions determine the stochastic variation of amino acid changes along a lineage. This Markovian process furnishes us with a fundamental statistical measure on possible genetic histories. Using this measure, we can understand evolution using tools from information geometry and statistical mechanics.
Mutational clock and statistical measure
A central concern of my research is building precise statistical descriptions of genotypic variations from foundational notions. The replication mistakes along a lineage describe a so called molecular clock, one which ticks differently for different kinds of mistakes. This Markovian process plays a role similar to the phase space of classical mechanics: it sets the base statistical measure of genotypic variation in an evolving population. It is the raw material that Darwinian evolution acts upon. I’m fascinated by the bridge between the microscopic gene dynamics and pathogenic evolution that we observe in disease.